Page added 22 October 2018 by Julia Koehler Leman.
Scientific Benchmarks are tests that compare Rosetta generated structure predictions with experimental observations. Assessing the accuracy of Rosetta predictions will
- Inform researchers on the trustworthiness of Rosetta as a molecular modeler
- Guide tuning Rosetta to improve structural predictions
Scientific benchmarks are meant to measure the physical realism of the energy function and how well a protocol is at sampling physically realistic structures.
How to set up a scientific benchmark test
Scientific test directory structure
Several tests are located in the Rosetta/main/tests directory. The directory structure is the following:
-
Rosetta/main/tests/integrationcontains integration tests -
Rosetta/main/tests/benchmarkcontains files required for the benchmark test server, i.e. the framework which runs the scientific tests, you might have to look them over when debugging -
Rosetta/main/tests/scientificcontains the scientific tests -
Rosetta/main/tests/scientific/testscontains the implementations of the tests, one directory per test -
Rosetta/main/tests/scientific/datasubmodule that contains the input data if >5 MB per test -
Rosetta/main/tests/scientific/tests/_template_template directory with all necessary input files
Where and how to add your test
- To set up a scientific benchmark, you will need to think about
- what your input data are
- you will want to have a diverse benchmark set that is realistic and not just structures that give you the best possible output
- the command line you are running
- make sure you are running either a baseline protocol (for instance de novo structure prediction, ddg_monomer, etc) that has been run by many people for many years or a state-of-the-art protocol: since these tests are computationally expensive, we need to ensure what we are testing makes sense
- what the output data look like
- in many cases we analyze score files but you might want to look at something different
- what your quality measures are
- are the quality measures you are using reasonable? Are there better measures in existence?
- what cutoffs you want to define to make the test a pass/fail
- you should have a general idea when you run the test locally but if you don’t know the specific cutoffs yet, don’t worry; you can set them later (see below)
- however, you want to think about HOW you define a pass/fail. It is encouraged to define it with respect to cutoffs that you define, i.e. running a single, self-contained test defines the initial cutoffs. We don’t prohibit cutoffs that are defined over time, for example running it 10x and comparing to previous runs (similar to regression tests), but this behavior is not necessarily encouraged.
- many tests evaluate the quality of folding funnels (plots of energy vs. RMSD to native), and can use already-developed tools for evaluating these that are insensitive to stochastic changes in the position of points, such as the PNear metric (see note below).
- the entire test should not run more than 1000-2000 CPU hours
- the runtime depends on the number of structures you run the benchmark on, the number of output models, and the protocol runtime per output model
- what your input data are
-
cd Rosetta/main/tests/scientific - run
git submodule update --init --recursiveto get the submodule containing the input data. You will now see aRosetta/main/tests/scientific/datadirectory - if your input data are larger than 5 MB, create a directory in the data submodule directory where you drop your input files, otherwise you can keep them in the tests directory (see below)
- commit your changes in that submodule
- move out of the data directory (
cd ..) and commit your changes again - [during that process: if you use zsh or check for
git statusyou will notice when git complains about uncommitted files]
-
cd Rosetta/main/tests/scientific/tests - copy the contents of the template directory to create your own scientific test directory via
cp -r Rosetta/main/tests/scientific/tests/_template_ <my_awesome_test> -
cd <my_awesome_test>: tests are set up by individual steps numbered sequentially so that they can be run individually without having to rerun the entire pipeline. Don’t run anything yet (we’ll get to that further below). - Put together your test, don’t run anything yet: edit each file in this directory wherever you find the
==> EDIT HEREtags.- if you are not running a RosettaScripts protocol, you can delete the fast_relax.xml file
-
0.compile.py: this script is for compilation, likely you won’t need to edit this file -
1.submit.py: this contains the command line and the target proteins you are running. The debug option does not refer to the release vs debug run in Rosetta but rather to the debug version of running the scientific test for faster setup. -
2.analyze.py: this script analyzes the results from the score files or whatever files you are interested in. It reads the cutoffs file and compares this run’s results against them. It will write theresult.txtfile for easy reading. A few basic functions for data analysis are at the bottom of this script. If you need your specialized function, please add it there. -
3.plot.py: this script will plot the results via matplotlib intoplot_results.pngwith subplots for each protein. It draws vertical and horizontal lines for the cutoffs. -
9.finalize.py: this script gathers all the information from the readme, the results plot and the results and creates a html pageindex.htmlthat displays everything - add all relevant papers to the
citationfile - edit the
cutoffsfile, header line starts with#and has one protein per row, different measures in columns - make sure you add your email to the
observers - fill out all the sections in the
readme.mdin as much detail as you can. Keep in mind that after you have left your lab, someone else in the community should be able to understand, run and maintain your test! This is automatically linked to the Gollum wiki. - add your test to the
runfunction at the bottom ofRosetta/main/tests/benchmark/tests/scientific/command.pyto hook it into the test server framework - a couple of notes on the test:
- we aim for plots to show the data, not just tables. Please include tables and/or the data in
jsonformat, but plots are highly encouraged as digesting data visually is faster and easier for us for debugging. - we like
jsonformat because it’s easy to read the data directly into python dictionaries. - a test should ideally be self-contained to obtain the cutoffs. As described above, try to avoid regression-test-like behavior where you compare to previous results or gather results from multiple runs over time.
- be consistent with the test name, keep a single name that is also the name of the test directory. It appears in multiple files and consistency makes debugging easier
- we aim for plots to show the data, not just tables. Please include tables and/or the data in
- Install python3.6 or later if you haven’t already, anything earlier won’t work. Make sure you have an alias set where you an specify the python version, for instance
python3. Every python script in the scientific tests will need thepython3prefix to run them properly!
Run your tests locally
-
Setup a run on Multiple Cores
- If you want to run locally using multiple cores, copy
main/tests/benchmark/benchmark.ini.templatetobenchmark.linux.ini(or whatever your architecture is). Adjust the settings in this file (i.e.cpu_countandmemory) as appropriate for your environment. Ifhpc_driver = MultiCore, this will submit jobs up tocpu_countwithout using an HPC job distributor.
- If you want to run locally using multiple cores, copy
-
Setup the run
-
cd Rosetta/main/tests/benchmark*python3 benchmark.py --compiler <clang or else> --skip \--debug scientific.<my_awesome_test>- the
--skipflag is to skip compilation, only recommended if you have an up-to-date version of master compiled in release mode (Sergey advises against skipping) - the
--debugflag is to run in debug mode which is highly recommended for debugging (i.e. you create 2 decoys instead of 1000s)
- the
- this creates a directory
Rosetta/main/tests/benchmark/results/<os>.scientific.<my_awesome_test>where it creates softlinks to the files inRosetta/main/tests/scientific/tests/<my_awesome_test>and then it will likely crash in one way or another
-
-
Start Debugging
- for step-by-step debugging
cd Rosetta/main/tests/benchmark/results/<os>.scientific.<my_awesome_test>and debug each script individually, starting from the lowest number, by running for instancepython3 1.submit.py - note that several other files and folders are created in the process:
-
config.jsonwhich contains the configuration settings - an
outputdirectory is created that contains the subdirectories for each protein - an
hpc-logsdirectory is created that contains the Rosetta run logs. You might have to check them out to debug your run if it crashed in the Rosetta run step. - each numbered script will also have an associated
.jsonfile that contains the variables you want to carry over into the next step -
9.finalize.output.jsoncontains all the variables and results saved -
plot_results.pngwith the results -
index.htmlwith the gathered results, failures and details you have written up in the readme. While all the files are accessible on the test server later, this file is the results summary that people will look at -
output.results.jsonwill tell you whether the tests passed or failed
-
- for step-by-step debugging
Submit your branch
-
Once you are finished debugging locally, commit all of your changes to your branch
- create a pull-request
- run the test on the test server
- since scientific tests require a huge amount of computational time, you might want to lower your
nstructfor debugging your run on the test server. If you do that, don’t forget to increase it later once the tests run successfully
- since scientific tests require a huge amount of computational time, you might want to lower your
-
Once the tests run as you want, merge your branch into
master- The
scientificbranch is an extra branch that grabs the latest master version every few weeks to run all scientific tests on. DO NOT MERGE YOUR BRANCH INTO THE SCIENTIFIC BRANCH!!! - tell Sergey Lyskov (sergey.lyskov@gmail.com) that your test is ready to be continuously run on the scientific branch
Celebrate! Congrats, you have added a new scientific test and contributed to Rosetta’s greatness. :D
- The
A common problem: evaluating folding funnel quality
Frequently, a scientific test will aim to evaluate the quality of a folding funnel (a plot of Rosetta energy vs. RMSD to a native or designed structure). Many of the simpler ways of doing this suffer from the effects of stochastic changes to the sampling: the motion of a single sample can drastically alter the goodness-of-funnel metric. For example, one common approach is to divide the funnel into a "native" region (with an RMSD below some threshold value) and a "non-native" region (with an RMSD above the threshold), and to ask whether there is a large difference between the lowest energy in the "native" region and the lowest in the "non-native" region. A single low-energy point that drifts across the threshold from the "native" region to the "non-native" region can convert a high-quality funnel into a low one, by this metric.
To this end, the PNear metric was developed. PNear is an estimate of the Boltzmann-weighted probability of finding a system in or near its native state, with "native-ness" being defined fuzzily rather than with a hard cutoff. The expression for PNear is:
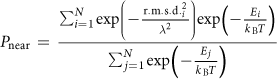
Intuitively, the denominator is the partition function, while the numerator is the sum of the Boltzmann probability of individual samples multiplied by a weighting factor for the "native-ness" of each sample that falls off as a Gaussian with RMSD. The expression takes two parameters: lambda (λ), which determines the breadth of the Gaussian for "native-ness" (with higher values allowing a more permissive notion what is close to native), and kB*T, which determines how high energies translate into probabilities (with higher values allowing states with small energy gaps to be considered to be closer in probability). Recommended values are lambda = 2 to 4, kB*T = 1.0 (for ref2015) or 0.63 (for talaris2013).
For more information, see the Methods (online) of Bhardwaj, Mulligan, Bahl et al. (2016). Nature 538(7625):329-35.
Update: As of 10 October 2019, a Python script is available in the tools repository (in tools/analysis) to compute PNear. Instructions for its use are in the comments at the start of the script. In addition, the function calculate_pnear(), in Rosetta/main/tests/benchmark/util/quality_measures.py, can be used to compute PNear.
The science behind your test: Scientific test template
Please use this template to describe your scientific test in the readme.md as described above. Also check out the fast_relax test for ideas of what we are looking for.
## AUTHOR AND DATE #### Who set up the benchmark? Please add name, email, PI, month and year
## PURPOSE OF THE TEST #### What does the benchmark test and why?
## BENCHMARK DATASET #### How many proteins are in the set? #### What dataset are you using? Is it published? If yes, please add a citation. #### What are the input files? How were the they created?
## PROTOCOL #### State and briefly describe the protocol. #### Is there a publication that describes the protocol? #### How many CPU hours does this benchmark take approximately?
## PERFORMANCE METRICS #### What are the performance metrics used and why were they chosen? #### How do you define a pass/fail for this test? #### How were any cutoffs defined?
## KEY RESULTS #### What is the baseline to compare things to - experimental data or a previous Rosetta protocol? #### Describe outliers in the dataset.
## DEFINITIONS AND COMMENTS #### State anything you think is important for someone else to replicate your results.
## LIMITATIONS #### What are the limitations of the benchmark? Consider dataset, quality measures, protocol etc. #### How could the benchmark be improved? #### What goals should be hit to make this a "good" benchmark?